Research Focus
We take an interdisciplinary approach that combines proteomics technology with biological inquiries
Spatiotemporal proteomics
Understanding the subcellular regulations of protein homeostasis
Many diseases are caused by changes in proteins inside the cell. These changes can come from altering the degree to which a gene is expressed by a cell type, making more or fewer copies of a protein. It is increasingly recognized however, that the level of expression of a gene is far from the only way cells control proteins. For instance, proteins can get damaged when they perform their function, and some proteins are more prone to damage. Ensuring that old proteins are effectively recycled over time, i.e., protein turnover, is critical to maintaining cellular activity. In parallel, where inside the cell a protein can be found, i.e., its subcellular location, can have profound influence on its functionality. Our lab is interested in developing subcellular spatial proteomics methods to investigate protein localization and translocation under stress and disease conditions, and to combine spatial proteomics methods with protein turnover measurements to compare proteostatic statuses across subcellular locales. Some of our ongoing work include:
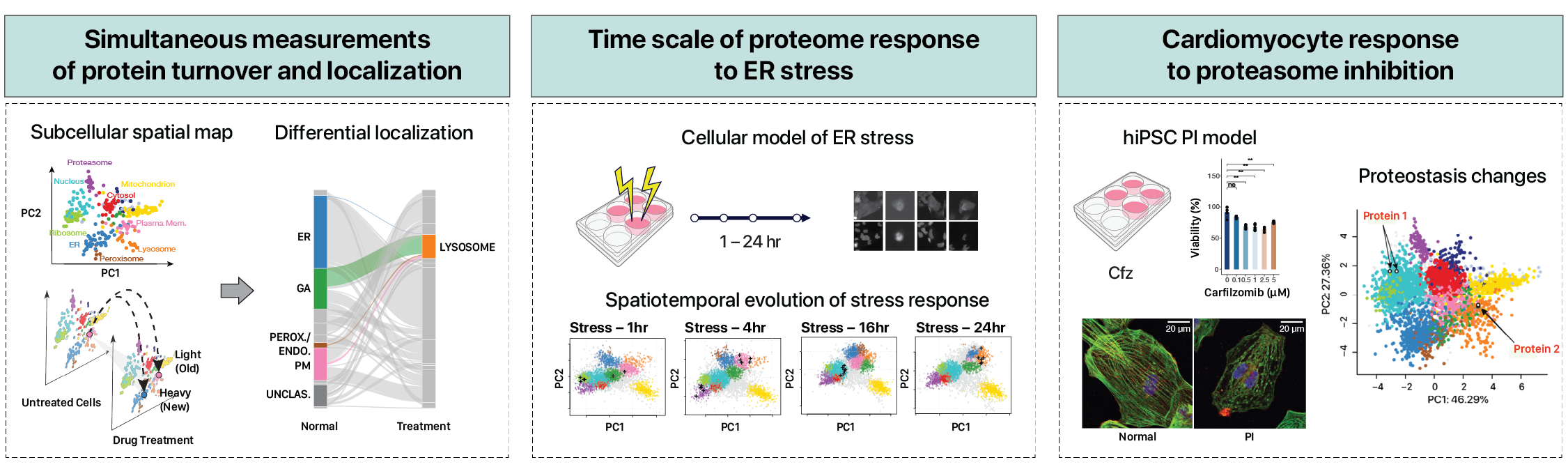
Simultaneous Proteome Localization and Turnover Analysis
Different compartments within the cell have distinct protein degradation mechanisms responsible for recycling their resident proteins. One of the research focuses of our lab is develop new methods that can measure spatial and temporal proteomic changes in the same samples. To reveal the the dynamics of proteins over space and time, we developed a new experimental approach that combines spatial and temporal protein tagging. The first isotope label tags proteins with isotopes to record the way they segregate under high speed ultracentrifugation, which allows us to deduce where inside the cell a protein is found. Using a different type of isotope label, we can also measure the rates that heavier elements appear over time in the isolated proteins, as a way to calculate the rate at which cells are recycling proteins. Coupled to mass spectrometry measurements, the results revealed that under cellular stress and cancer drug toxicity, cells changes in protein turnover rates according to which organelles they are found. At the same time, our methods are revealing a growing list of proteins that are spatially regulated -- in other words, proteins that translocate or move from their original sub cellular location to a new locale.
Effect of the cancer drug carfilzomib on cardiac cells
We are interested in applying spatiotemporal proteomics to find out why some commonly prescribed cancer drugs may be toxic to the heart of certain patients. Using human induced pluripotent stem cell (iPSC) models of human heart cells in a dish, we found that surprisingly the cancer drug carfilzomib may disrupt the protein organization in the human heart. On the other hand, carfilzomib may specifically block the turnover of certain proteins that work together to generate the contractile force of the heart. This finding may help explain why many patients develop heart failure after cancer drug treatment, and may open up future strategies to reduce the level of toxicity.
Team Members
Related Publications
Protein Turnover Dynamics Analysis With Subcellular Spatial Resolution.
Alamillo, Lorena , Black, Alexander , Lam, Maggie P Y …
Bio Protoc (2025)Simultaneous proteome localization and turnover analysis reveals spatiotemporal features of protein homeostasis disruptions.
Currie, Jordan , Manda, Vyshnavi , Robinson, Sean K , …
Nat Commun (2024)Simultaneous proteome localization and turnover analysis reveals spatiotemporal features of protein homeostasis disruptions.
Currie, Jordan , Manda, Vyshnavi , Robinson, Sean K , …
bioRxiv (2024)Last updated: January 27, 2026


