Research Focus
We take an interdisciplinary approach that combines proteomics technology with biological inquiries
Protein turnover kinetics
We develop methods to measure protein synthesis and degradation flux
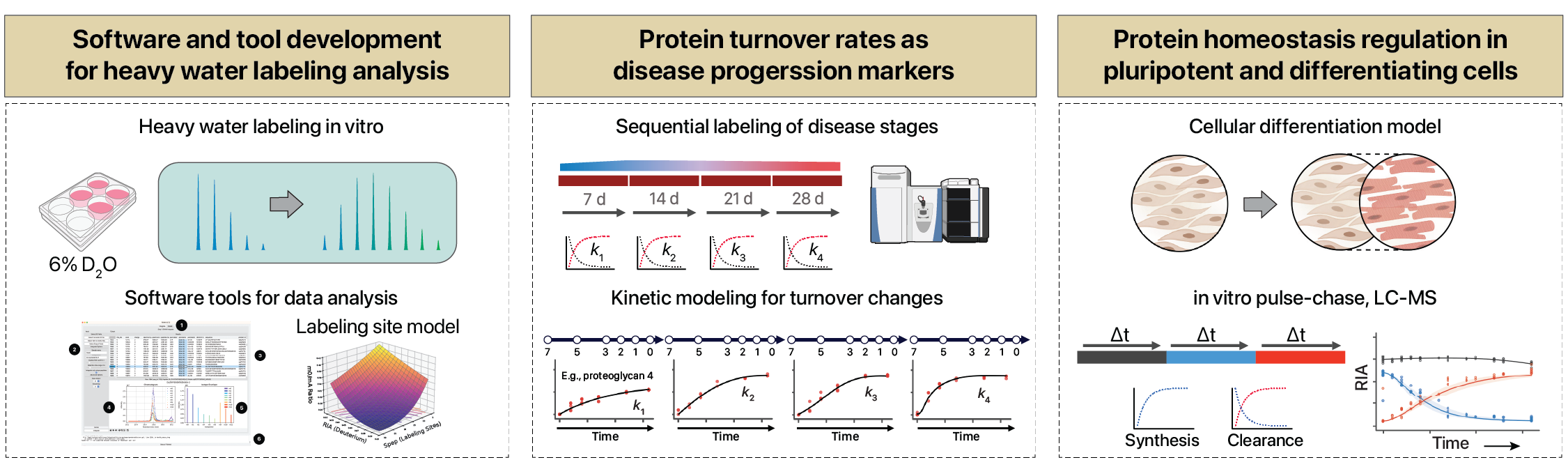
Protein turnover, or the continued synthesis and degradation of cellular proteins, is a fundamental feature of life and plays an important role in the regulation of gene expression. Moreover, protein turnover determines the trajectory of physiological processes such as growth, development, hypertrophy, and atrophy, as well as aging-related debilitations. Our lab is interested in developing experimental and analytical methods to determine protein half-life in cells and tissues. Some of our areas of focus include:
Methods to measure protein turnover rates in vivo using heavy water labeling
Mass spectrometry combined with stable isotope labeling can be used to measure protein turnover rates on a large scale. Turnover rates in adult animals can differ greatly from those in cell culture, and can range from minutes to decades. Measuring turnover rates in animals in vivo however faces additional challenges, including how best to introduce the stable isotope tracer, the delay in tracer availability across organs, and potential issues of label reutilization. In previous work, our team has used heavy water (D2O) to tag newly synthesized proteins with deuterium at stable, non-exchangeable hydrogens. D2O is an attractive tracer for mammalian protein turnover analysis because it has a good safety profile and also equilibrates quickly with heavy water. Our lab has applied this strategy to identify protein turnover changes during cardiac hypertrophy. More recently, we have worked with collaborators in the Leinwand Lab at UC Boulder to apply D2O labeling to compare sarcomere turnover in different muscle types. In ongoing work, we are applying D2O labeling to understand extracellular matrix protein synthesis and degradation in cardiac fibrosis.
Comparing of labeling and analytic strategies
Working with the Beynon Lab at the University of Liverpool, we are interested in controlled comparison of heavy water vs amino acid labeling for measuring protein turnover rates, and examining whether different labeling methods and analytical pipelines return comparable numerical values in protein turnover rates and half-life. Amino acid labeling is commonly used as a metabolic protein precursor in animal models to measure in vivo protein turnover rate, but there are usually delays in how fast the labeled amino acid precursors become available for protein synthesis. This difference is likely dependent on the metabolism of the tissue too, and will delay the label incorporation into the measured protein, especially for proteins with very short half-life. By comparing different kinetic models, and methods to derive the precursor kinetic parameters used to adjust half-life measurement, we are investigating best practices for correcting for precursor kinetics.
Software development for protein half-life calculation
Our lab created a Python software tool, Riana, which can be used to perform mass spectrometry data integration and kinetics curve fitting for both amino acid and heavy water studies. Ongoing work is enhancing the functionality and performance of Riana, and its applicability to different types of labeling study. In a recent work, we showed that an automated mass isotopomer selection method can lead to 20-50% gain in the depth of D2O labeling experiments, measured by the number of peptides that are well fitted to kinetic models. In a more recent work, we are extending D2O labeling to in vitro cell cultures and differentiating cells in non-equilibrium conditions.
Team Members
Related Publications
Deuterium labeling enables proteome-wide turnover kinetics analysis in cell culture.
Alamillo, Lorena , Ng, Dominic C M , Currie, Jordan …
Cell Rep Methods (2025)Improved Method to Determine Protein Turnover Rates with Heavy Water Labeling by Mass Isotopomer Ratio Selection.
Currie, Jordan , Ng, Dominic C M , Pandi, Boomathi …
J Proteome Res (2025)Deuterium labeling enables proteome wide turnover kinetics analysis in cell culture.
Alamillo, Lorena , Ng, Dominic C M , Currie, Jordan …
bioRxiv (2025)Improved determination of protein turnover rate with heavy water labeling by mass isotopomer ratio selection.
Currie, Jordan , Ng, Dominic C M , Pandi, Boomathi …
bioRxiv (2024)Harmonizing Labeling and Analytical Strategies to Obtain Protein Turnover Rates in Intact Adult Animals.
Hammond, Dean E , Simpson, Deborah M , Franco, Catarina …
Mol Cell Proteomics (2022)Integrated omics dissection of proteome dynamics during cardiac remodeling.
Lau, Edward , Cao, Quan , Lam, Maggie P Y …
Nat Commun (2018)A large dataset of protein dynamics in the mammalian heart proteome.
Lau, Edward , Cao, Quan , Ng, Dominic C M …
Sci Data (2016)Last updated: January 27, 2026




